Direct visualization of floppy 2D DNA origami using cryo-EM
- Journal
- iScience
- Volume
- 25
- Page
- 104373
- Year
- 2022
- Date
- 2022-06-01
Abstract
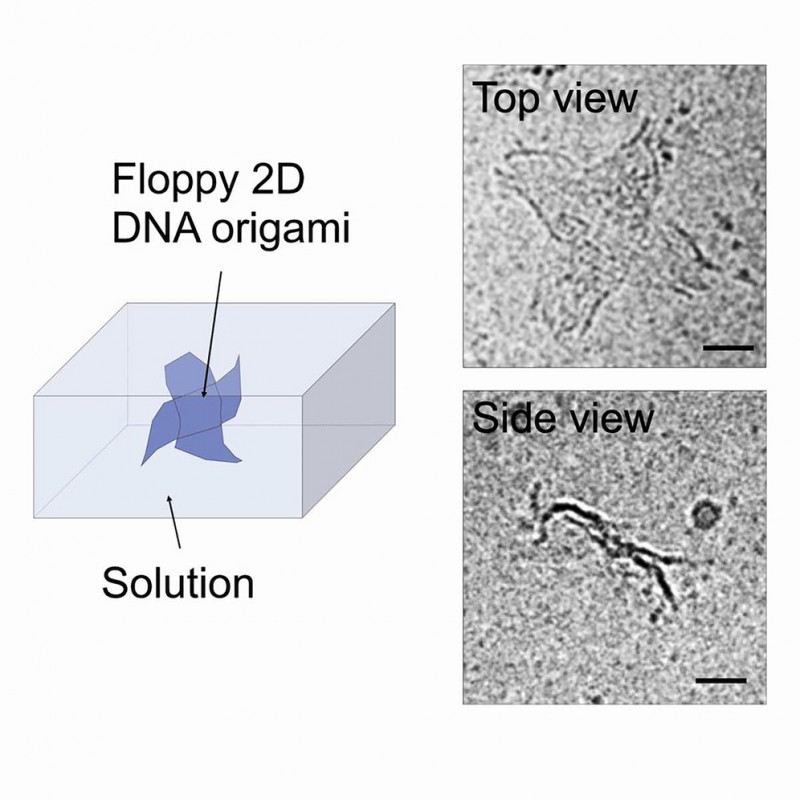
DNA origami enables self-assembly of small two-dimensional DNA origami tiles into customizable large-scale objects with designated structures, which hold the promise for various applications in nanotechnology and biotechnology. Since the morphology and properties of the DNA monomers in solution greatly influence their assembly processes, determining the true aqueous morphology and properties of DNA origami tiles in solution is key for further development in this emerging field, which, however, remains challenging. Here, we utilize cryo-EM to directly visualize the aqueous structure of DNA origami cross-tiles and their assemblies. By immobilizing the sample in amorphous ice, we observed the floppy morphology of DNA cross-tile monomers and the intermolecular contacts within larger assemblies. In addition, we observed the interactions between cross-tiles that trigger the formation of clusters and stacks of DNA cross-tiles in solution, which could potentially affect the assembly and folding of DNA origami. Finally, we calculated the structure and flexibility of DNA cross-tiles and their trimer assemblies using finite element analysis, providing insights into the distorted structure and self-folding of DNA origami in solution. Our discovery has laid the foundation for investigating the construction of large DNA assemblies in solution, informing knowledge regarding the self-replication and self-folding mechanisms of 2D DNA origami.